A New Way to Discover the Structures of Membrane Proteins
University of Toronto scientists have discovered a better way to extract proteins from the membranes that encase them, making it easier to study how cells communicate with each other to create human health and disease.
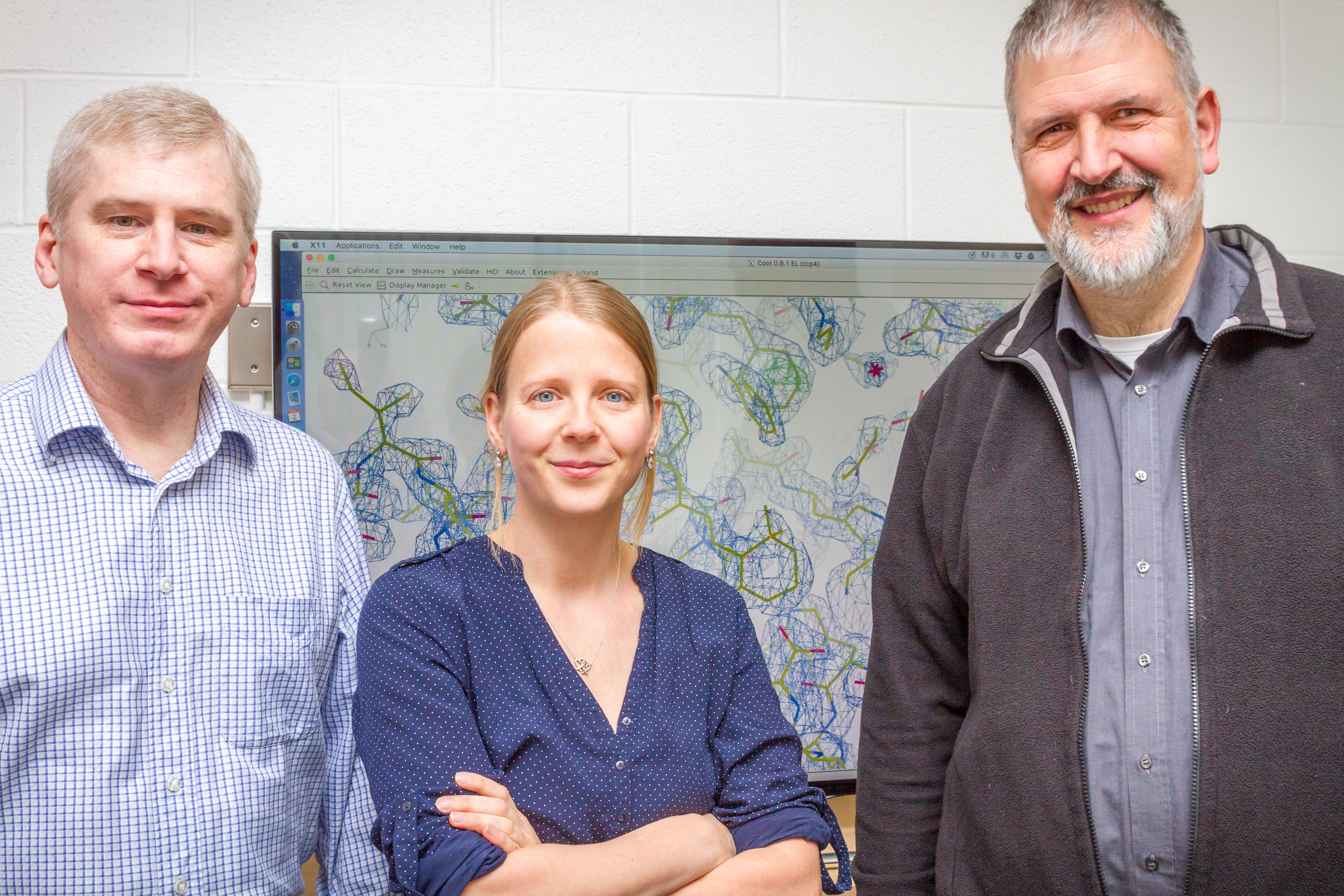
Scientists are very interested in understanding how membrane proteins work and why they malfunction under certain circumstances. Looking at their 3D structures is a particularly useful way to do this. Currently, researchers use detergents to separate proteins from their fatty membrane casing for further in-detail studies. But detergent strips the fat molecules away from the proteins, which very often destabilizes the proteins and makes them useless for study. As a result, new 3D structures are rarely discovered and published.
Dr. Jana Broecker, a postdoctoral fellow in the lab of Professor Oliver P. Ernst, discovered that she could use a type of plastic, or polymer, originally developed by the auto industry, to better stabilize these crucial proteins and thereby make them available for 3D structure determination.
The polymers don’t strip off fat molecules from the proteins, instead wrapping themselves around the protein, with the fat molecules still attached, says Broecker, of the Department of Biochemistry. Using the new substance, she was able to keep these necessary fat molecules attached while she studied the protein’s 3D structure using X-ray crystallography.
“We believe this approach can be applied to many more membrane proteins, which would drastically speed up structure discovery of currently unknown membrane proteins,” says Broecker. “With more and better structures at hand, it will be easier to develop new drugs for the treatment of human diseases in the near future.”
Broecker and colleagues published their findings Feb. 7 on the cover of the journal Structure.

Optimize this page for search engines by customizing the Meta Title and Meta Description fields.
Use the Google Search Result Preview Tool to test different content ideas.
Select a Meta Image to tell a social media platform what image to use when sharing.
If blank, different social platforms like LinkedIn will randomly select an image on the page to appear on shared posts.
Posts with images generally perform better on social media so it is worth selecting an engaging image.
Heidi Singer
Unpacking the Genetics of Inflammation
 One in five people have a genetic change that’s associated with worse arthritis and sepsis, a serious bacterial infection. University of Toronto researcher Tania Watts has discovered how this tiny change worsens these conditions by increasing inflammation in our bodies.
One in five people have a genetic change that’s associated with worse arthritis and sepsis, a serious bacterial infection. University of Toronto researcher Tania Watts has discovered how this tiny change worsens these conditions by increasing inflammation in our bodies.
Watts first published her results in the online edition of Nature Immunology Nov. 28.
For those who have the genetic trait, each infection they get makes a little less of an anti-inflammatory protein called TRAF1, which is normally created in white blood cells during infections. This deficiency causes increased inflammation each time someone has an infection and makes inflammatory diseases like rheumatoid arthritis a bit worse.
Although this genetic trait does not cause disease on its own, in people who have the constellation of traits that lead to rheumatoid arthritis, it appears that people with this particular genetic trait are more likely to develop disease or have worse disease symptoms.
“Although we don’t know the full implications yet, it is possible that people with rheumatoid arthritis who have this genetic change may need to be monitored more carefully for flare-ups after an infection and treated accordingly,” says Watts, a professor of Immunology in the Faculty of Medicine. “It might be more important to treat infections more quickly and aggressively for these patients.”
The genetic change is probably a leftover from evolutionary days, as inflammation is important in the control of infection. However, the downside is that for people with chronic, inflammatory diseases, this trait makes inflammation worse -- leading researchers to examine its role in more detail.
“Understanding how one gene is contributing to the disease, knowing how each genetic contribution is modifying the immune response we hope will be useful for designing more customized therapies,” says Watts.

Optimize this page for search engines by customizing the Meta Title and Meta Description fields.
Use the Google Search Result Preview Tool to test different content ideas.

Select a Meta Image to tell a social media platform what image to use when sharing.
If blank, different social platforms like LinkedIn will randomly select an image on the page to appear on shared posts.
Posts with images generally perform better on social media so it is worth selecting an engaging image.
Heidi Singer

U of T Scientists Co-Discover How to Turn Off CRISPR
CRISPR genome editing is quickly revolutionizing biomedical research, but the new technology is not yet exact. The technique can inadvertently make excessive or unwanted changes in the genome and create off-target mutations, limiting safety and efficacy.

Now, researchers at the University of Toronto and University of Massachusetts Medical School have discovered the first known “off-switches” for CRISPR gene editing activity, providing much greater control over the edits, according to a new study featured on the cover of Cell.
The scientists found three proteins that block CRISPR, known as anti-CRISPRs.
Professor Alan Davidson, of the Departments of Molecular Genetics and Biochemistry, and Assistant Professor Karen Maxwell of the Department of Biochemistry, made the discovery with UMass researcher Erik J. Sontheimer.
“CRISPR is very powerful, but we have to be able to turn it off,” says Davidson. “This is a very fundamental addition to the toolbox which should give researchers more confidence to use gene editing.”
A simple and efficient way of editing the genome, CRISPR is changing biomedical research by making it far easier to inactivate or edit genes in a cell line for study. Work that used to take months or years to perform can now be done in weeks.
Scientists are developing CRISPR to target specific cell types, tissues or organs where a disease occurs. But sometimes, CRISPR hits the wrong target, causing unintended damage.
“CRISPR activity in these other cells, tissues or organs is at best useless and at worst a safety risk,” says Sontheimer. “But if you could build an off-switch that keeps Cas9 (the enzyme that cuts the DNA for editing) inactive everywhere except the intended target tissue, then the tissue specificity will be improved.”
The new paper not only identifies that “off switch” but it shows that CRISPR inhibitors have evolved naturally and can be identified and exploited.
The “off switch” will allow researchers to be more precise in their use of CRISPR. If they only want to use it during one stage of a cell’s life – such as when the DNA is replicating – they can turn it off during all other stages, reducing the chance of unwanted consequences.
A major way of delivering CRISPR into the body is through inactivated viruses that can be programmed to attach themselves to target cells. The challenge is that viruses can’t be engineered to be 100 per cent specific.

Researchers in muscular dystrophy, for example, want to target muscle cells. But a particular virus known for its ability to target muscle cells also attaches itself to liver cells, where it could cause unintended damage. The “off switch” could allow researchers to release “anti-CRISPR” proteins into the body to turn off CRISPR activity in liver cells, offering a new layer of protection against mistakes.
“Knowing we have a safety valve will allow people to develop many more uses for CRISPR,” says Maxwell. “Things that may have been too risky previously might be possible now.”
The “off switch” could be used across the board for any application of CRISPR technology to target specific cells or tissues. For the researchers, the next step is to widen the “off switch” to include other types of CRISPR systems.

Optimize this page for search engines by customizing the Meta Title and Meta Description fields.
Use the Google Search Result Preview Tool to test different content ideas.

Select a Meta Image to tell a social media platform what image to use when sharing.
If blank, different social platforms like LinkedIn will randomly select an image on the page to appear on shared posts.
Posts with images generally perform better on social media so it is worth selecting an engaging image.
Heidi Singer

Seeing Double: Twins Combine Passion for Rehabilitation and Dance
Twin sisters Lindsey and Alison Schwartz share a combined love of dance and travel. They will have a chance to combine the two when they travel to the Immortelle Centre for Special Education in Trinidad to provide rehabilitation services and dance classes to students with special needs. Lindsey, a physical therapy (PT) student, and Alison, an occupational therapy (OT) student, are participating in an inter-professional clinical placement that will allow the sisters to work side by side, helping the patients enhance their coordination and physical function. Hosted by the University of Toronto’s International Centre for Disability and Rehabilitation (ICDR), this internship provides physical and occupational therapy students with a unique opportunity to gain clinical experience, while expanding their understanding of disability and rehabilitation within a global context. Writer Alyson Musial sat down with Lindsey and Alison to discuss their upcoming journey.
What inspired your interests in rehabilitation?
L: We both grew up as competitive dancers, and visited physical therapists throughout our dance careers. Our mom is an occupational therapist, so rehabilitation was always a topic discussed in our home. In high school and university, we volunteered at the Hospital for Sick Children. We worked on different units, sparking our individual interests. When we both got accepted at University of Toronto, we were so happy and knew it was the right choice.
A: Studying rehabilitation in parallel with Lindsey is really helpful. As we progress in our programs, we discuss what we are learning, which lets us think with a more interdisciplinary mind. When faced with an OT challenge, I often find myself analyzing from both a PT and OT perspective. I think, “How would Lindsey approach this, and how can I use that different analysis to enhance my OT approach”.
What drew you to the ICDR-Trinidad internship?
A: I was attracted to the internship because the Immortelle Centre for Special Education has a long affiliation with University of Toronto, spanning over 15 years. ICDR-Trinidad is a team of faculty, clinicians and students at U of T, and they helped us speak with students who went in years past; everyone had such positive things to say, we knew we needed to pursue this opportunity. It also allows PTs and OTs to work together, which doesn’t always happen in clinical internships.
L: Ali and I both have an interest in working with children when we graduate, so it seemed like a great fit. Also, international placements aren’t as common for PTs, so I jumped at the chance to broaden my horizons. This placement offers a lot of independent and collaborative work, which will allow us to build our critical thinking and planning skills, as well as our clinical confidence.
Tell us about the new dance program you will be organizing for the students.
A: We have always wanted to combine our passions for dance and rehabilitation. Lindsey and I thought the clinical internship offered by ICDR-Trinidad would be a great chance to develop a program that lets children experience the therapeutic aspects of dance. We have been told that the students at Immortelle love to dance and move, and that dance is a big part of their culture.
L: I think dancing with the students will also help us build a trusting relationship with them. The program will take place during their school day, and our goal is to plan a performance for their Christmas show, giving the students something to progress towards. We want them to experience the joy of dance.
A: In the future, Lindsey and I would like to create a rehabilitation-focused dance program for kids with special needs, so this opportunity is a perfect start. We can’t wait!

Optimize this page for search engines by customizing the Meta Title and Meta Description fields.
Use the Google Search Result Preview Tool to test different content ideas.

Select a Meta Image to tell a social media platform what image to use when sharing.
If blank, different social platforms like LinkedIn will randomly select an image on the page to appear on shared posts.
Posts with images generally perform better on social media so it is worth selecting an engaging image.
Toronto Emerges as Big Data Hotspot
“Information is the new oil,” say University of Toronto scientists as city becomes one of the world’s leading producers of biodata

University of Toronto (U of T) announced today that it is launching a massive project to sequence the whole genomes of 10,000 people per year starting in 2016. The project accelerates the big data revolution, positioning Toronto as a leader in the global race to understand complex diseases and create a new generation of precision therapies.
“With sequencing now married to large-scale computation, I believe we in Toronto can help lead the way for precision medicine,” said Professor Stephen Scherer, director of U of T’s McLaughlin Centre and the Centre for Applied Genomics at The Hospital for Sick Children (SickKids). “Genomic information is the new oil. It’s the resource that’s going to drive technology in the new era. We’re generating the oil that researchers will use to enable discoveries and to create new products in software, biotechnology and information management that will realize precision medicine.”
In 2015, Toronto scientists, led by Professor Scherer, earned competitive Canada Foundation for Innovation funding to purchase the Illumina HiSeq X™ Ten sequencing system, along with powerful computers that together can process the 10,000 genomes a year. Previously, Toronto researchers had been forced to send genomes to offshore labs for sequencing, causing research delays and significantly higher costs. The project will generate data from thousands of control genomes serving as crucial controls for the world’s researchers. The genomes, to be sequenced in Scherer’s lab, will provide medical researchers the necessary yardstick by which to compare disease.
“The groundbreaking work that U of T and SickKids have done for the MSSNG Autism Genome Sequencing Project and the Canadian Personal Genome Project has set the stage for the creation of a large pediatric reference genome that will have far reaching utility in complex disease,” said Christian Henry, Executive Vice President and Chief Commercial Officer of Illumina. “We believe Toronto’s research community is already linked to vast repositories of patient health data and is committed to finding answers in complex diseases.”
Toronto's whole genome project is working with leading health tech innovators, including Verily Life Sciences, an Alphabet company.
“Whole genome sequencing is bringing the worlds of life science and data science together,” said David Glazer, on the Steering Committee for the Global Alliance for Genomics and Health, founder of Google Genomics, and engineering director at Verily Life Sciences. “The combination of those two disciplines is one of the most exciting frontiers in medicine today. Our existing collaboration with Steve's lab on the 10,000-genome MSSNG project is a good example of the opportunity; we're eager to see the many more such projects enabled by today's announcement.”
Big Data, the massive streams of information now being generated through advances in computational power, is transforming medical research. Whole-genome sequencing is used to understand life at its most fundamental level but also to find unique genetic defects that underlie disease in individuals, and design diagnostics and precision treatments for them.
“Toronto has unique advantages in the world that allow us to generate high-quality data sets and manage those securely and ethically,” said Professor Trevor Young, Dean of the Faculty of Medicine at the University of Toronto, which encompasses nearly 6,000 faculty members from 33 hospitals in the Greater Toronto Area. “Within a mile radius we have one of the world’s leading medical schools, connected to most of Canada’s top hospitals and other key research fields such as computer science and engineering, as well as the headquarters of Canada’s top financial institutions.”
Precision medicine will come about when genetic data is matched to other kinds of health and lifestyle information. In Toronto, a single medical school connects all the city’s hospitals to each other and to other key research fields including the social and physical sciences. Meanwhile, university researchers, physicians and hospitals are connected to the provincial government and to the patients of Toronto through the single-payer health insurance system. The population of Toronto is also among the most diverse in the world, providing an unusual breadth of genetic material to study.
For many years, Ontario researchers have analyzed anonymized patient health information through the single payer system, and Toronto scientists such as Lap-Chee Tsui, who identified the genetic defect causing cystic fibrosis, have been at the forefront of genetic research. Now, Toronto researchers have the ability to produce massive streams of genomic data at a reasonable cost.

Optimize this page for search engines by customizing the Meta Title and Meta Description fields.
Use the Google Search Result Preview Tool to test different content ideas.

Select a Meta Image to tell a social media platform what image to use when sharing.
If blank, different social platforms like LinkedIn will randomly select an image on the page to appear on shared posts.
Posts with images generally perform better on social media so it is worth selecting an engaging image.
Heidi Singer

Sport, Disability and Media: How Are the Paralympic Games Covered?
With the 2016 Paralympic Games rapidly approaching in Rio de Janeiro from September 7 to 18, writer Alyson Musial sat down to chat with Nancy Quinn, a sport physiotherapist and University of Toronto Masters graduate who researches the intersection of sport, disability and media. Nancy is a veteran of six Paralympic Games and has received a Queen Elizabeth II Diamond Jubilee Medal for her contribution to the Paralympic movement in Canada. Alyson also spoke to Nancy’s advisor, Department of Physical Therapy Professor Karen Yoshida, about how Paralympic athletes are represented in the media.

Nancy, tell us why you are so passionate about the Paralympic Games.
Nancy: Where to start? I graduated in 1987 with a BScPT fully entrenched in the medical model of disability; patients with a disability had a problem that required fixing. I had spent 4 years learning the biology and pathology of impairment, and had been taught strategies and techniques to repair, modify and at the very least, limit the progress of impairment. Upon reflection, I received little if any exposure to people outside of the acute care setting who lived with physical difference. In four years of university I met no one who worked outside the home, had a family, played sport, dated, and lived with a disability.
And so began my illustrious career of fixing people who I really knew nothing about.
A series of fortunate circumstances placed me on the medical team of Team Canada at the 1996 Paralympic Games in Atlanta, Georgia. Here I met a community of people with diverse physical impairments, who were highly athletic, and were passionate about elite sport and competition. These same people travelled, danced, dined out, had children, married and divorced (not necessarily in that order), worked, and lived with a disability. These people also faced a variety of societal challenges.
Post Atlanta, I was hooked. I had discovered a sporting community where physical difference did not preclude athletic excellence. I returned home to central Ontario brimming with stories and enthusiasm to discover that there had been no media coverage of these Games whatsoever.
Zero.
Nothing in The Globe and Mail, our national newspaper, and no television coverage. My friends and family congratulated my volunteerism with these Games, using language of charity and benevolence. How good it was of me to help those people out! I found this very frustrating. Six Paralympic Games later and I am a proud and passionate advocate for the Canadian Paralympic movement and a disability scholar in the making.
How are disabled athletes represented in the Canadian media?
Karen: Sport journalism has traditionally featured athletes with disability far less often than able-bodied athletes. When athletes with a disability do receive coverage, their disability is framed as a tragedy, and that sport offers the athlete the opportunity to recover from, or transcend, this tragedy to achieve more “normal”, able-bodied life. This framework celebrates their athletic achievement, but only as a triumph over the personal tragedy of impairment. Also, female Paralympic athletes are represented less often than their male counterparts. Male athletes who use wheelchairs, who are white and identify as heterosexual receive more frequent, diverse coverage than other athletes with disability.
Our research also shows that Paralympic athletes are faced with a complex dilemma: when the sport media choose to represent a Paralympian as an elite athlete by minimizing their physical difference, the athlete becomes more relatable to a non-disabled viewing audience. Yet conversely, embracing physical difference establishes credibility for athletes within their disabled community. The tension between minimizing difference for non-disabled audiences, and highlighting difference for audiences with a disability, is a very interesting and challenging reality for the Paralympic community and makers of media.
How do conversations around disability, sport and the media affect rehabilitation?
Karen: It’s no mystery that media plays an influential role in our lives. Rehabilitation professionals need to work diligently to think critically about disability, which can be challenging given most clinicians are educated and socialized professionally in the biomedical model of disability, wherein disability equates to incapacity. Discussions of athleticism, and therefore ability, challenge assumptions of the biomedical model and encourage new ways of thinking about rehabilitation, which is great! As media continues to construct more alternative, positive cultural representations of athletes and ability, the language and practice around physical difference will evolve within the field of rehabilitation.
Are media representations of para-sport evolving?
Nancy: The good news is, things are improving. Working with Karen while doing my Masters, I looked at CBC’s coverage of the 2004 Paralympic Games. CBC did a great job of representing our Canadian Paralympic athletes as athlete first. This was a truly positive step. Since the early 2000s, there is evidence to support that media representations of female and male Paralympic athletes have been growing, in quantity and quality. Slowly, the media is embracing more multi-dimensional representations of Paralympic.
It’s 2016 and my inbox is full of great media regarding para-sport in Canada and the pending Rio Paralympic Games, informed by person first/athlete first language. I find videos on YouTube of people with physical difference dancing, giving advice on dating or how to prepare for a job interview, and playing sport.
But it’s 2016 and I still see people with mobility difference unable to move through the snow on a city sidewalk that has not been cleared. I hear able bodied people in my clinic waiting room speaking overly loud to others who have obvious physical difference. I see big budget films at the theater with story lines that reinforce the tragedy of disability.
It’s 2016 and media representation of para-sport, Paralympians and disability has evolved and continues to evolve. We’ve come a long way, but we have a long way to go.

Optimize this page for search engines by customizing the Meta Title and Meta Description fields.
Use the Google Search Result Preview Tool to test different content ideas.

Select a Meta Image to tell a social media platform what image to use when sharing.
If blank, different social platforms like LinkedIn will randomly select an image on the page to appear on shared posts.
Posts with images generally perform better on social media so it is worth selecting an engaging image.
A More Powerful Way to Develop Therapeutics?
A University of Toronto scientist has developed a new method for identifying the raw ingredients necessary to build ‘biologics’, a powerful class of medications that has revolutionized treatment of diseases like rheumatoid arthritis and some cancers.

Biologics are a type of drug that results from the high-tech manipulation of our own proteins, as opposed to more traditional drugs built from synthetic chemicals. Because of their success so far, scientists are racing to create new biologics – and now, a University of Toronto researcher has developed a way to make that process more powerful.
Philip M. Kim, an associate professor in U of T’s Donnelly Centre for Cellular and Biomolecular Research, combined high-tech computer simulation and high-throughput laboratory experiments to create what he hopes will be the most effective way to discover the proteins that are key to new biologics. His research was published online in the journal Science Advances on July 20, 2016.
“A large fraction of new therapeutics these days involve engineered proteins that latch onto a drug target, for instance on a cancer cell,” says Kim, also of the departments of Molecular Genetics and Computer Science. “Finding a protein that effectively binds to a target can feel like looking for a needle in a haystack. Our method should open up new opportunities to find those key proteins – and make a major impact on the development of new biologics.”
Under the traditional approach to developing a biologic, researchers identify a protein of interest and then test billions of variants, either randomly generated or from a natural source, hoping to find an effective binder. But these methods allow very little control over where and how the protein performs this crucial function on its target – a major factor in its effectiveness.
Kim and his team took a different approach. They used a computer to simulate the binding process, and then designed proteins that would work on the target. This type of theoretical approach has been in development for several decades, but is still not effective enough. So Kim combined the best of both methods. Instead of randomly creating massive libraries of variants, as with the traditional approach, he used computer modelling to generate a smaller, but intelligently designed repertoire of variants. Designing each variant allows for the tight control of all its properties, in contrast to conventional approaches.
“We showed that this method gives you binders that are somewhat stronger than what you get with the conventional approach,” says Kim. “The much smaller library also solves many technical problems, and we can screen for new, previously unscreenable, targets. It’s a very exciting time for cancer research, and for biologics.”
For Kim, the next step is to produce proteins that are important to certain types of cancer, but have not been screened before due to the difficulty producing them.

Optimize this page for search engines by customizing the Meta Title and Meta Description fields.
Use the Google Search Result Preview Tool to test different content ideas.

Select a Meta Image to tell a social media platform what image to use when sharing.
If blank, different social platforms like LinkedIn will randomly select an image on the page to appear on shared posts.
Posts with images generally perform better on social media so it is worth selecting an engaging image.
Heidi Singer

A Better Way to Measure the Severity of Prostate Cancer?
A University of Toronto researcher has found that the key to understanding the severity of prostate cancer may be in the urine.

Professor Thomas Kislinger, of the Department of Medical Biophysics, and his co-authors have made a major advance in the quest to develop a precise, non-invasive method of identifying which tumors are severe enough to require urgent and aggressive treatment.
Overtreating prostate cancer is a worldwide problem because many of these cancers are slow growing, and may never require treatment, says Kislinger, who is also Senior Scientist at the Princess Margaret Cancer Centre, University Health Network.
“We believe we have found a better way that allows us to predict which patients have a slow-growing versus aggressive prostate cancer using non-invasive biomarkers,” he says. “This could eventually help us personalize cancer treatment for these patients.”
The research was published online June 29 in Nature Communications. Kislinger discusses details at https://youtu.be/dj6LBuXYyoo.
Currently, needle biopsies are used to help diagnose prostate cancer. But this technique may not detect hidden tumors, or cancer that has already spread beyond the organ, according the authors.
The Kislinger team, along with Canadian and American collaborators, used urine samples containing prostatic secretions from 210 patients.
The research took four years and involved samples from 300 patients, says Kislinger, who specializes in proteomics – the study of the structure and function of proteins involved in the development of cancer.
“We used targeted proteomics to accurately quantify hundreds of proteins in urine samples to identify liquid biopsy signatures. The first round of research involved 80 patients and quantified 150 proteins that were then narrowed down to 34 for further investigation. The next round involved a second, independent cohort of 210 patients,” he says.
“Applying computational biology, we used the quantitative data from mass spectrometry to develop the fluid biomarkers for aggressive prostate cancer.”
Next, the team intends to study urine samples from 1,000 international patients to determine whether the biomarkers identified have broader clinical utilities in prostate cancer.

Optimize this page for search engines by customizing the Meta Title and Meta Description fields.
Use the Google Search Result Preview Tool to test different content ideas.

Select a Meta Image to tell a social media platform what image to use when sharing.
If blank, different social platforms like LinkedIn will randomly select an image on the page to appear on shared posts.
Posts with images generally perform better on social media so it is worth selecting an engaging image.

A Better Way to Predict Diabetes
University of Toronto and Kaiser Permanente scientists develop a highly accurate method to predict type 2 diabetes after delivery in women with gestational diabetes

An international team of researchers has discovered a simple, accurate new way to predict which women with gestational diabetes will develop type 2 diabetes after delivery. The discovery would allow health care providers to identify women at greatest risk and help motivate women to make early lifestyle changes and follow other strategies that could prevent them from developing the disease later in life.
Gestational diabetes is defined as glucose intolerance that is first identified during pregnancy. It occurs in three to 13 percent of all pregnant women, and increases a woman’s risk of developing type 2 diabetes by 20 to 50 percent within five years after pregnancy.
The joint efforts of the University of Toronto’s Michael Wheeler, a professor in the Department of Physiology, and Erica Gunderson, Senior Research Scientist with the Kaiser Permanente Northern California Division of Research, led to development of a technique called targeted metabolomics to better predict the development of type 2 diabetes in women with recent gestational diabetes. Typically, diabetes is diagnosed by measuring blood sugar levels in the form of glucose, an important fuel used by cells in the body. The researchers identified several other metabolites that indicate early changes that signify future diabetes risk long before changes in glucose levels occur.
The team tested fasting blood samples collected from women with gestational diabetes within two months after delivery — predicting with 83 percent accuracy which women would develop the disease later on. These results were significantly better at predicting the development of type 2 diabetes than conventional methods, a fasting blood test followed by the time-consuming and inconvenient oral glucose tolerance test.
“After delivering a baby, many women may find it very difficult to schedule two hours for another glucose test,” says Wheeler, who is also a Senior Scientist at the Toronto General Research Institute. “What if we could create a much more effective test that could be given to women while they’re still in the hospital? Once diabetes has developed, it’s very difficult to reverse.”
“Early prevention is the key to minimizing the devastating effects of diabetes on health outcomes,” says Dr. Gunderson. “By identifying women soon after delivery, we can focus our resources on those at greatest risk who may benefit most from concerted early prevention efforts.”
The fasting blood samples used for this study were obtained from 1,035 women diagnosed with gestational diabetes and enrolled in the Kaiser Permanente’s Study of Women, Infant Feeding and Type 2 Diabetes after GDM Pregnancy, also known as the SWIFT Study, which was funded by the U.S. National Institutes of Health (R01 HD050625). The SWIFT study screened women with oral glucose tolerance tests at 2 months after delivery and then annually thereafter to evaluate the impact of breastfeeding and other characteristics on the development of type 2 diabetes after a pregnancy complicated by gestational diabetes.
The American Diabetes Association recommends type 2 diabetes screening at six to 12 weeks after delivery in women with gestational diabetes, and every one to three years afterwards for life. The time-consuming nature of the two-hour oral glucose test is believed to be one reason for low compliance rates of less than 40 percent in some settings.
The new method may also be able to predict individuals who may develop type 2 diabetes in the general population – a major advance at a time when more than 300 million people suffer from the preventable form of this disease. A next-generation blood test that’s more simple and accurate than the current options could help to identify individuals who would benefit most from more timely and effective interventions to prevent type 2 diabetes.
Wheeler and Gunderson are now hoping to conduct additional tests in women with gestational diabetes to evaluate racial and ethnic differences in prediction, and investigate high risk groups with prediabetes to learn if metabolomics will predict type 2 diabetes in the general population.

Optimize this page for search engines by customizing the Meta Title and Meta Description fields.
Use the Google Search Result Preview Tool to test different content ideas.

Select a Meta Image to tell a social media platform what image to use when sharing.
If blank, different social platforms like LinkedIn will randomly select an image on the page to appear on shared posts.
Posts with images generally perform better on social media so it is worth selecting an engaging image.
Heidi Singer

Blood Test Identifies women at Risk of Preterm Delivery as Early as 17 Weeks of Pregnancy
A blood test developed by University of Toronto and international researchers has been shown to predict if a pregnant woman is at risk of delivering her baby prematurely, before the full 37 weeks of gestation.

At 18 weeks, the test can predict premature delivery with 82 per cent accuracy. At 28 weeks, the accuracy rises to 86 per cent.
Premature birth remains the main cause of child-related mortality in the developed world. It occurs in five to 10 per cent of all pregnancies, but is associated with 70 per cent of all newborn deaths (excluding genetic anomalies) and up to 75 per cent of newborn disease including cerebral palsy, blindness, deafness, respiratory illness and complications of neonatal intensive care.
The study was published on June 22, 2016 in PLOS one, and was led by Dr. Stephen Lye, a professor in the departments of Obstetrics and Gynaecology and Physiology, along with researchers from Harvard University and the University of Calgary.
“There are treatments that can prevent preterm birth,” say Lye, who is also a Senior Scientist at the Lunenfeld-Tanenbaum Research Institute, part of the Sinai Health System. “But these treatments are only useful in a subset of women. This blood test could improve identification of women who will benefit from existing therapies. Moreover, it may also help drug studies to focus on women who are at highest risk of delivering preterm when evaluating new treatments.”
The study population is a subset of women who participated in the All Our Babies study, a community based longitudinal pregnancy cohort in Calgary, Alberta. The researchers collected paired maternal blood from pregnant women at two clinically relevant time points: approximately 17 weeks when fetal ultrasound is conducted and at approximately 27 weeks of gestation when gestational diabetes screening is performed.
The international team, consisting of clinicians, scientists and biostatisticians, used gene expression profiling and bioinformatics to develop gene sets, coupled with a patient’s clinical information such as history of preterm birth, history of abortion or anaemia, to predict whether or not a woman will deliver prematurely.

Optimize this page for search engines by customizing the Meta Title and Meta Description fields.
Use the Google Search Result Preview Tool to test different content ideas.

Select a Meta Image to tell a social media platform what image to use when sharing.
If blank, different social platforms like LinkedIn will randomly select an image on the page to appear on shared posts.
Posts with images generally perform better on social media so it is worth selecting an engaging image.
